Home
Long non-coding RNAs (lncRNAs) are endogenous cellular RNAs that are mRNA-like in length (> 200 nt) but which lack any positive-strand open-reading frames longer than 30 amino acids. LncRNAs are increasingly implicated in infectious disease, however, only a few have been functionally characterized for their role during viral infection.
Using the 8 founder mice of the Collaborative Cross (http://compgen.unc.edu/), we quantified the expression of 20,728 mouse lncRNA genes, 5,329 of which were differentially expressed after influenza A virus (IAV) or severe acute respiratory syndrome coronavirus (SARS-CoV) infection. We annotated these differentially expressed lncRNAs by 2 computational methods (module-based and rank-based enrichments).
Using MONOCLdb, researchers can retrieve and visualize annotations, expression profiles and functional enrichment results of specific long-non coding RNAs. On-the-fly generation of expression heatmaps or generation of functional enrichment results (module-based enrichments, rank-based enrichments, associated GO term, associated IMMGEN module, associated REACTOME pathways) are possible using MONOCLdb. Co-expression networks and genomic networks of lncRNAs can also be visualized using MONOCLdb. Association scores between lncRNAs and pathogenicity variables are also provided by MONOCLdb. MONOCLdb also provides a Distributed Annotation System (DAS) service for visualization purposes of the non-coding genes associated with inferred biological functions (associated GO term, IMMGEN module, REACTOME pathways). Furthermore, MONOCLdb offers a web service to automatically retrieve analysis results (please see the About section for more information).
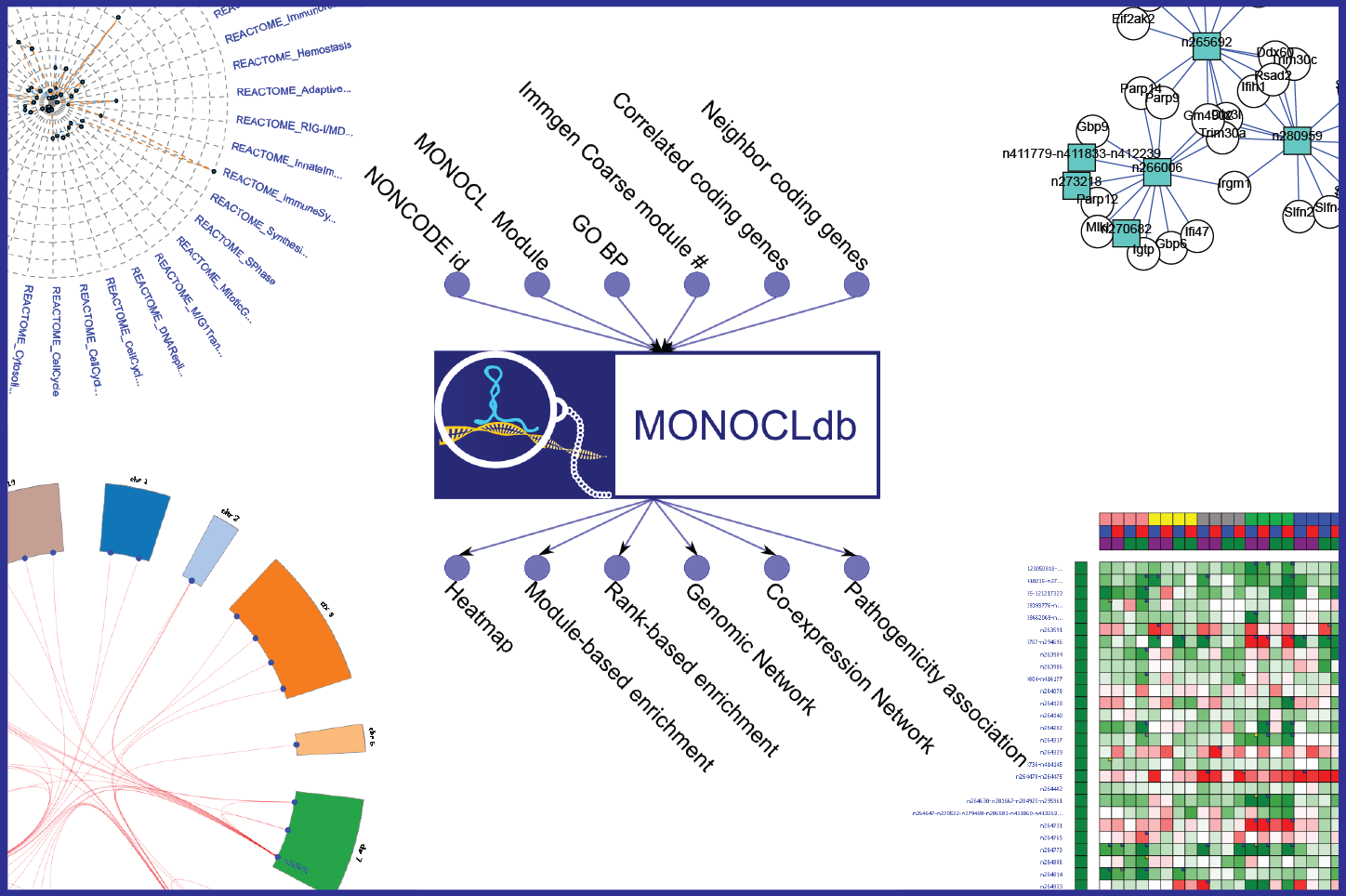
(1) Select lncRNAs by: NONCODE v3.0 identifiers (e.g. “n424068”), GO term found significantly enriched with the ranked-based annotation (e.g. “GO:0007010”), Immgen Coarse module number found significantly enriched with the ranked-based annotation (e.g. “Immgen_Coarse.module_28”), Ensembl gene ID of most correlated coding-genes (e.g. “ENSMUSG00000029088”), or Ensembl gene ID of chromosomic neighbor (within 200 kb) coding-genes (e.g. “ENSMUSG00000030921”).
(2) Submit the query.
(3) Visualize and retrieve the figures and annotations using the different tab menus.