Home
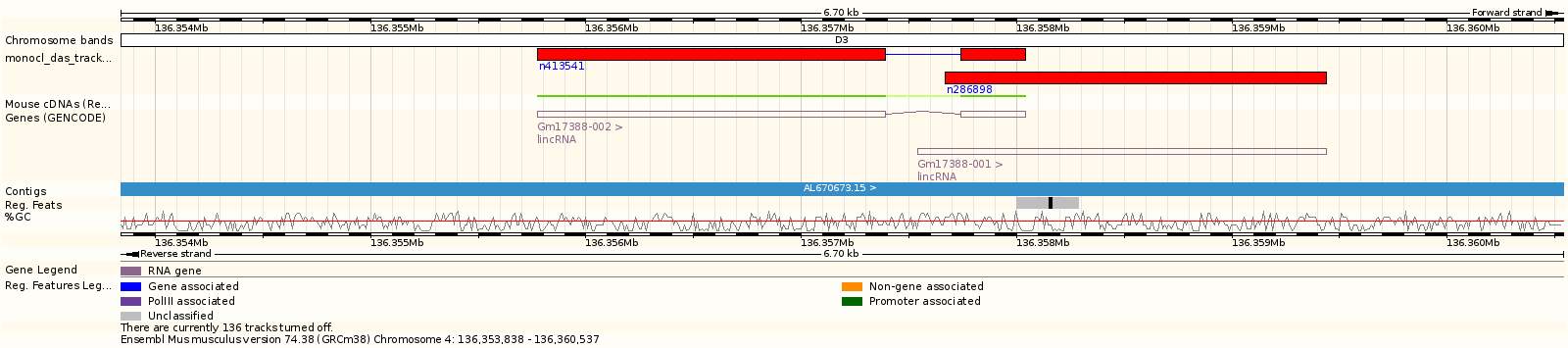
DAS allows providing annotation information of genomics data into a large variety of Genome Browsers (Ensembl, NCBI, UCSC, etc...). Further information regarding DAS can be found at http://www.dasregistry.org/ and http://www.biodas.org/
Please use https://www.monocldb.org:9000/das/ as DAS entry point for the MONOCL database. MONOCLdb DAS status:

The monocl DAS track can be used within all DAS-compliant Genome Browsers.
Note: You can directly access to the MONOCLdb DAS track on the reference Mouse genome using the Ensembl Genome Browser using this link: http://uswest.ensembl.org/Mus_musculus/Share/4364585da442eaba21b340b599eaa1d01526182